
A discussion of these details can be found in other publications.Įach of the various programs in the BLAST suite accepts a large number of options try running blastn -help to see them for the blastn program. Scoring matrices representing these rule sets with names like BLOSUM and PAM have been developed using a variety of methods to capture these considerations. But for dissimilar species separated by vast evolutionary time, such a mismatch might not be as bad relative to other possibilities. When comparing protein sequences from two similar species, for example, we might wish to give a poor score to the relatively unlikely match of a nonpolar valine (V) to a polar tyrosine (Y). Finally, rpsblast searches for sequence matches against sets of profiles, each representing a collection of sequences (as in HMMER, though not based on hidden Markov models).Īll this talk of scoring rules indicates that the specific scoring rules are important, especially when comparing two protein sequences. The deltablast program considers a precomputed database of scoring rules for different types of commonly found (conserved) sequences. This process is repeated as many times as the user wishes, with more dissimilar matches being revealed in later iterations. Other more exotic BLAST tools include psiblast, which produces an initial search and tweaks the scoring rules on the basis of the results these tweaked scoring rules are used in a second search, generally finding even more matches. The tblastx program compares nucleotide queries against nucleotide subjects, but it does so in protein space with all six conversions compared to all six on both sides. Generally such programs result in six times as much work to be done. The blastx and tblastn programs do this by converting nucleotide sequences into protein sequences in all six reading frames (three on the forward DNA strand and three on the reverse) and comparing against all of them. While two nucleotide sequences (N comparisons in the figure above) may be compared directly (as may two protein sequences, represented by P), when we wish to compare a nucleotide sequence to a protein sequence, we need to consider which reading frame of the nucleotide sequence corresponds to a protein. Depending on what type the query and subject sets are, different BLAST programs are used. The programs in the BLAST+ suite can search for and against sequences in protein format (as we did for the HMMER example) and in nucleotide format (A’s, C’s, T’s, and G’s).
#Blast 2 sequences manual#
The NCBI manual covers quite a few powerful and handy features of BLAST on the command line that this book does not. Reading the help information (e.g., with blastn -help) and the NCBI BLAST Command Line Applications User Manual at is highly recommended. This chapter only briefly covers running BLAST on the command line in simple ways. The most modern version of the software, called BLAST+, is maintained by the National Center for Biotechnology Information (NCBI) and may be downloaded in binary and source forms at. For example, an E value of 0.05 means that we can expect a match by chance in 1 in 20 similar searches, whereas an E value of 2.0 means we can expect 2 matches by chance for each similar search.īLAST is not a single tool, but rather a suite of tools (and the suite grows over the years as more features and related tools are added).
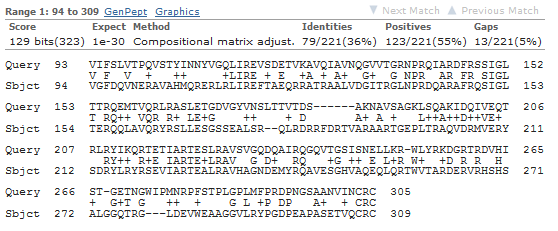
Because in larger subject sets some good matches are likely to be found by chance, each HSP is also associated with an “ E value,” representing the expected number of matches one might find by chance in a subject set of that size with that score or better. Each HSP is associated with a “bitscore” that is based on the similarity of the subsequences as determined by a particular set of rules. Sometimes, however, the term “hit” is used loosely, without differentiating between the two. A sufficiently close match between subsequences (denoted by arrows in the figure above, though matches are usually longer than illustrated here) is called a high-scoring pair (HSP), while a query sequence is said to hit a target sequence if they share one or more HSPs.


 0 kommentar(er)
0 kommentar(er)
